Make a phylogeny from species names alone
rOpenSci package: taxize
There is only functionality for plants right now in taxize. We hope to add more taxan groups soon.
Load libraries
library("taxize")
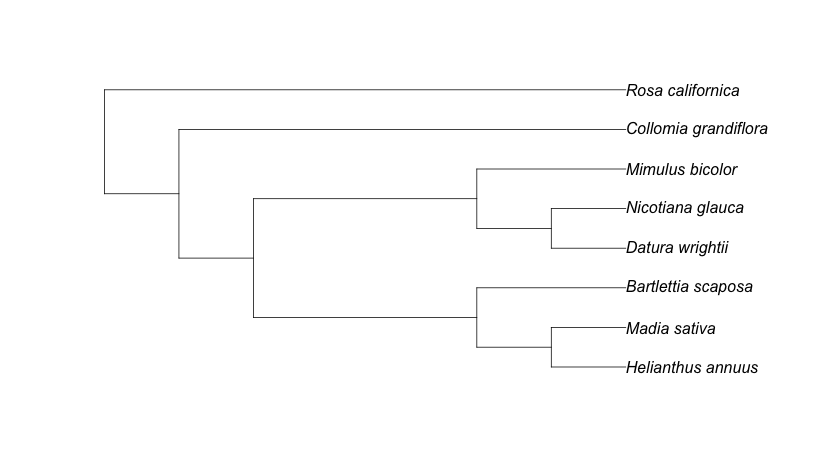
splist <- c("Helianthus annuus", "Collomia grandiflora", "Rosa californica", "Datura wrightii", "Mimulus bicolor", "Nicotiana glauca","Madia sativa", "Bartlettia scaposa")
Phylomatic is a web service with an API that we can use to get a phylogeny. Fetch phylogeny from phylomatic
phylogeny <- phylomatic_tree(splist)
Format tip-labels
phylogeny$tip.label <- capwords(phylogeny$tip.label, onlyfirst = TRUE)
Plot phylogeny
plot(phylogeny)