rvertnet tutorial
for v0.6.2
rvertnet is a client for interacting with VertNet.org. VertNet is a online database of vertebrate specimens/observations. They have a really nice web interface at http://portal.vertnet.org/search, but of course to do reproducible research you may want to use this package.
This package retrieves data, makes maps, and summarizes data from VertNet, with functions for searching by many parameters, including taxonomic names, places, dates, and more. In addition, there is an interface for conducting spatially delimited searches, and another for requesting large datasets via email.
Installation
You can install the stable version from CRAN:
install.packages("rvertnet")
Or the development version from GitHub using the devtools package:
if (!require("devtools")) install.packages("devtools")
devtools::install_github("ropensci/rvertnet")
library('rvertnet')
Usage
Search by term
Search for Aves in the state of California, limit to 10 records
res <- searchbyterm(class = "Aves", state = "California", limit = 10, verbose = FALSE)
All major functions (searchbyterm(), spatialsearch(), vertsearch()) give back a meta (for metadata, in a list) and data (for data, in a data.frame) slot. The metadata:
res$meta
#> $request_date
#> [1] "2017-10-17T19:54:47.475780"
#>
#> $response_records
#> [1] 10
#>
#> $submitted_query
#> [1] "class:Aves stateprovince:California"
#>
#> $request_origin
#> [1] "45.523452,-122.676207"
#>
#> $limit
#> [1] 10
#>
#> $last_cursor
#> [1] "False:Cu4GCskECpwE9wAAABn_____jIGJmo2LkZqL0o-QjYuek96WkZuah9LNz87M0s_H0s_H_wAA_3RtoKCZi4ygoP8AAP9dno-PmpGYlpGa_wAA_3N0bZaRm5qH_wAA_12biJz_AAD_c3Rtm5CcoJab_wAA_12cipKJ0J2WjZvSjI-anJaSmpGM0JeLi4_Sno2ci5CM0puei56dnoya0pKKjJqKktKYipab0pyKkonSnZaNm9LLy87JydKMmpab0s3Mz8zMxsj_AAD_c3-cipKJ0J2WjZvSjI-anJaSmpGM0JeLi4_Sno2ci5CM0puei56dnoya0pKKjJqKktKYipab0pyKkonSnZaNm9LLy87JydKMmpab0s3Mz8zMxsj_AAD__wD-__6MgYmajYuRmovSj5CNi56T3paRm5qH0s3PzszSz8fSz8f_AHRtoKCZi4ygoP8AXZ6Pj5qRmJaRmv8Ac3RtlpGbmof_AF2biJz_AHN0bZuQnKCWm_8AXZyKkonQnZaNm9KMj5qclpKakYzQl4uLj9KejZyLkIzSm56Lnp2ejJrSkoqMmoqS0piKlpvSnIqSidKdlo2b0svLzsnJ0oyalpvSzczPzMzGyP8Ac3-cipKJ0J2WjZvSjI-anJaSmpGM0JeLi4_Sno2ci5CM0puei56dnoya0pKKjJqKktKYipab0pyKkonSnZaNm9LLy87JydKMmpab0s3Mz8zMxsj_AP_-EAohBN0EkB08Gxk5AAAAAOb___9IClAAWgsJ4h5N-19FzJ8QAmDjpfACEg1Eb2N1bWVudEluZGV4Gu4BKEFORCAoSVMgImN1c3RvbWVyX25hbWUiICJhcHBlbmdpbmUiKSAoSVMgImdyb3VwX25hbWUiICJzfnZlcnRuZXQtcG9ydGFsIikgKElTICJuYW1lc3BhY2UiICJpbmRleC0yMDEzLTA4LTA4IikgKElTICJpbmRleF9uYW1lIiAiZHdjIikgKEFORCAoT1IgKFFUICJBdmVzIiAicnRleHRfY2xhc3MiKSAoSVMgInJhdG9tX2NsYXNzIiAiYXZlcyIpKSAoUVQgIkNhbGlmb3JuaWEiICJydGV4dF9zdGF0ZXByb3ZpbmNlIikpKToZCgwoTiBvcmRlcl9pZCkQARkAAAAAAADw_0oFCABA6Ac"
#>
#> $query_version
#> [1] "search.py 2016-08-15T16:43+02:00"
#>
#> $matching_records
#> [1] ">10000"
#>
#> $api_version
#> [1] "api.py 2017-01-12T20:08-03:00"
The data
res$data
#> # A tibble: 10 x 70
#> higherclassification
#> <chr>
#> 1 Animalia | Chordata | | | Strigidae | Otus
#> 2 Animalia | Chordata | | | Laridae | Sterna
#> 3 Animalia | Chordata | | | Recurvirostridae | Himantopus
#> 4 Animalia; Chordata; Aves; Passeriformes; Emberizidae;
#> 5 Animalia; Chordata; Aves; Passeriformes; Emberizidae; Ridgway, 1876
#> 6 Animalia; Chordata; Aves; Passeriformes; Emberizidae; Ridgway, 1876
#> 7 Animalia; Chordata; Aves; Anseriformes; Anatidae; Amazonetta brasiliensis
#> 8 Animalia; Chordata; Aves; Apodiformes; Trochilidae;
#> 9 Animalia; Chordata; Aves; Passeriformes; Emberizidae; Emberizinae;
#> 10 Animalia; Chordata; Aves; Passeriformes; Emberizidae; Emberizinae;
#> # ... with 69 more variables: stateprovince <chr>, basisofrecord <chr>,
#> # month <chr>, decimallongitude <chr>, phylum <chr>, references <chr>,
#> # year <chr>, startdayofyear <chr>, taxonrank <chr>,
#> # specificepithet <chr>, bibliographiccitation <chr>, family <chr>,
#> # countrycode <chr>, geodeticdatum <chr>,
#> # coordinateuncertaintyinmeters <chr>, highergeography <chr>,
#> # continent <chr>, verbatimlocality <chr>, day <chr>, kingdom <chr>,
#> # collectioncode <chr>, occurrencestatus <chr>,
#> # coordinateprecision <chr>, institutioncode <chr>,
#> # scientificname <chr>, locality <chr>, class <chr>,
#> # vernacularname <chr>, county <chr>, decimallatitude <chr>,
#> # occurrenceid <chr>, language <chr>, license <chr>, country <chr>,
#> # georeferenceverificationstatus <chr>, modified <chr>, eventdate <chr>,
#> # nomenclaturalcode <chr>, verbatimeventdate <chr>, genus <chr>,
#> # order <chr>, catalognumber <chr>, enddayofyear <chr>,
#> # locationremarks <chr>, infraspecificepithet <chr>, accessrights <chr>,
#> # identificationverificationstatus <chr>, identificationqualifier <chr>,
#> # occurrenceremarks <chr>, institutionid <chr>,
#> # georeferenceprotocol <chr>, georeferenceddate <chr>,
#> # georeferencedby <chr>, organismid <chr>, preparations <chr>,
#> # recordedby <chr>, individualcount <chr>, georeferencesources <chr>,
#> # dateidentified <chr>, previousidentifications <chr>,
#> # locationaccordingto <chr>, othercatalognumbers <chr>,
#> # identifiedby <chr>, associatedmedia <chr>, sex <chr>,
#> # dynamicproperties <chr>, verbatimcoordinatesystem <chr>,
#> # samplingprotocol <chr>, verbatimcoordinates <chr>
Search for Mustela nigripes in the states of Wyoming or South Dakota, limit to 20 records
res <- searchbyterm(specificepithet = "nigripes", genus = "Mustela", state = "(wyoming OR south dakota)", limit = 20, verbose = FALSE)
res$data
#> # A tibble: 19 x 73
#> month decimallongitude startdayofyear
#> <chr> <chr> <chr>
#> 1 12 -100.8276541162 336
#> 2 03 -100.9827 64
#> 3 1 -100.759483 1
#> 4 3 -100.73 67
#> 5 11 <NA> 305
#> 6 10 <NA> 282
#> 7 8 <NA> 234
#> 8 12 <NA> 342
#> 9 12 <NA> 358
#> 10 1 <NA> 1
#> 11 11 <NA> 313
#> 12 9 <NA> 272
#> 13 12 <NA> 335
#> 14 9 <NA> 259
#> 15 10 <NA> 297
#> 16 12 <NA> 339
#> 17 11 <NA> 305
#> 18 11 <NA> 315
#> 19 <NA> <NA> <NA>
#> # ... with 70 more variables: accessrights <chr>, kingdom <chr>,
#> # verbatimcoordinatesystem <chr>, day <chr>,
#> # identificationverificationstatus <chr>, occurrenceid <chr>,
#> # identificationqualifier <chr>, phylum <chr>, verbatimeventdate <chr>,
#> # coordinateuncertaintyinmeters <chr>, higherclassification <chr>,
#> # sex <chr>, year <chr>, specificepithet <chr>, basisofrecord <chr>,
#> # geodeticdatum <chr>, occurrenceremarks <chr>, highergeography <chr>,
#> # continent <chr>, scientificname <chr>, language <chr>,
#> # institutionid <chr>, country <chr>, genus <chr>,
#> # georeferenceprotocol <chr>, family <chr>, stateprovince <chr>,
#> # county <chr>, georeferenceddate <chr>, references <chr>,
#> # georeferencedby <chr>, verbatimlocality <chr>, institutioncode <chr>,
#> # organismid <chr>, eventtime <chr>, preparations <chr>,
#> # recordedby <chr>, license <chr>, dynamicproperties <chr>,
#> # georeferenceverificationstatus <chr>, modified <chr>, eventdate <chr>,
#> # individualcount <chr>, bibliographiccitation <chr>,
#> # verbatimcoordinates <chr>, georeferencesources <chr>,
#> # catalognumber <chr>, locationaccordingto <chr>, collectioncode <chr>,
#> # class <chr>, previousidentifications <chr>, decimallatitude <chr>,
#> # locality <chr>, othercatalognumbers <chr>, identifiedby <chr>,
#> # nomenclaturalcode <chr>, order <chr>, enddayofyear <chr>,
#> # minimumelevationinmeters <chr>, maximumelevationinmeters <chr>,
#> # samplingprotocol <chr>, dateidentified <chr>, eventremarks <chr>,
#> # datasetname <chr>, locationremarks <chr>, taxonrank <chr>,
#> # countrycode <chr>, occurrencestatus <chr>, vernacularname <chr>,
#> # recordnumber <chr>
Search for class Aves, in the state of Nevada, with a coordinate uncertainty range (in meters) of less than 25 meters
res <- searchbyterm(class = "Aves", stateprovince = "Nevada", error = "<25", verbose = FALSE)
res$data
#> # A tibble: 8 x 70
#> month decimallongitude startdayofyear minimumelevationinmeters
#> <chr> <chr> <chr> <chr>
#> 1 10 -119.582 288 1780
#> 2 10 -119.582 288 1780
#> 3 10 -119.582 288 1780
#> 4 10 -119.582 288 1780
#> 5 10 -119.582 288 1780
#> 6 10 -119.582 288 1780
#> 7 06 -114.09658 165 2072.64
#> 8 09 -118.57885 248 1786.128
#> # ... with 66 more variables: accessrights <chr>, kingdom <chr>,
#> # verbatimcoordinatesystem <chr>, day <chr>,
#> # identificationverificationstatus <chr>, occurrenceid <chr>,
#> # identificationqualifier <chr>, phylum <chr>, verbatimeventdate <chr>,
#> # coordinateuncertaintyinmeters <chr>, higherclassification <chr>,
#> # lifestage <chr>, modified <chr>, year <chr>, specificepithet <chr>,
#> # basisofrecord <chr>, geodeticdatum <chr>, highergeography <chr>,
#> # continent <chr>, scientificname <chr>, catalognumber <chr>,
#> # language <chr>, institutionid <chr>, country <chr>, genus <chr>,
#> # georeferenceprotocol <chr>, family <chr>, stateprovince <chr>,
#> # county <chr>, georeferenceddate <chr>, references <chr>,
#> # georeferencedby <chr>, verbatimlocality <chr>, habitat <chr>,
#> # institutioncode <chr>, organismid <chr>,
#> # maximumelevationinmeters <chr>, preparations <chr>, recordedby <chr>,
#> # sex <chr>, dynamicproperties <chr>,
#> # georeferenceverificationstatus <chr>, infraspecificepithet <chr>,
#> # samplingprotocol <chr>, eventdate <chr>, individualcount <chr>,
#> # bibliographiccitation <chr>, verbatimcoordinates <chr>,
#> # georeferencesources <chr>, dateidentified <chr>,
#> # locationaccordingto <chr>, collectioncode <chr>, class <chr>,
#> # previousidentifications <chr>, decimallatitude <chr>, locality <chr>,
#> # othercatalognumbers <chr>, identifiedby <chr>,
#> # nomenclaturalcode <chr>, order <chr>, enddayofyear <chr>,
#> # license <chr>, associatedmedia <chr>, occurrenceremarks <chr>,
#> # recordnumber <chr>, collectionid <chr>
Spatial search
Spatial search service allows only to search on a point defined by latitude and longitude pair, with a radius (meters) from that point. All three parameters are required.
res <- spatialsearch(lat = 33.529, lon = -105.694, radius = 2000, limit = 10, verbose = FALSE)
res$data
#> # A tibble: 10 x 60
#> month decimallongitude startdayofyear minimumelevationinmeters
#> <chr> <chr> <chr> <chr>
#> 1 07 -105.68633 193 2182.368
#> 2 07 -105.705479 196 2023.872
#> 3 07 -105.705479 196 2023.872
#> 4 07 -105.705479 196 2023.872
#> 5 07 -105.705479 196 2023.872
#> 6 07 -105.705479 196 2023.872
#> 7 07 -105.705479 196 2023.872
#> 8 07 -105.705479 196 2023.872
#> 9 07 -105.705479 196 2023.872
#> 10 07 -105.705479 196 2023.872
#> # ... with 56 more variables: accessrights <chr>, kingdom <chr>,
#> # day <chr>, identificationverificationstatus <chr>, occurrenceid <chr>,
#> # identificationqualifier <chr>, phylum <chr>, verbatimeventdate <chr>,
#> # coordinateuncertaintyinmeters <chr>, higherclassification <chr>,
#> # sex <chr>, year <chr>, specificepithet <chr>, basisofrecord <chr>,
#> # geodeticdatum <chr>, occurrenceremarks <chr>, highergeography <chr>,
#> # continent <chr>, scientificname <chr>, language <chr>,
#> # institutionid <chr>, country <chr>, genus <chr>,
#> # georeferenceprotocol <chr>, family <chr>, stateprovince <chr>,
#> # county <chr>, georeferenceddate <chr>, references <chr>,
#> # georeferencedby <chr>, verbatimlocality <chr>, institutioncode <chr>,
#> # organismid <chr>, maximumelevationinmeters <chr>, preparations <chr>,
#> # recordedby <chr>, dynamicproperties <chr>,
#> # georeferenceverificationstatus <chr>, modified <chr>, eventdate <chr>,
#> # individualcount <chr>, bibliographiccitation <chr>,
#> # georeferencesources <chr>, catalognumber <chr>,
#> # locationaccordingto <chr>, recordnumber <chr>, class <chr>,
#> # previousidentifications <chr>, decimallatitude <chr>, locality <chr>,
#> # othercatalognumbers <chr>, identifiedby <chr>,
#> # nomenclaturalcode <chr>, enddayofyear <chr>, order <chr>,
#> # collectioncode <chr>
Global full text search
vertsearch() provides a simple full text search against all fields. For more info see the docs. An example:
res <- vertsearch(taxon = "aves", state = "california", limit = 10)
res$data
#> # A tibble: 10 x 60
#> higherclassification stateprovince
#> <chr> <chr>
#> 1 Animalia | Chordata | | | Strigidae | Otus California
#> 2 Animalia | Chordata | | | Laridae | Sterna California
#> 3 Animalia | Chordata | | | Recurvirostridae | Himantopus California
#> 4 Aves | Galliformes | Odontophoridae Washington
#> 5 Aves | Galliformes | Odontophoridae Washington
#> 6 Aves | Galliformes | Odontophoridae Washington
#> 7 Aves | Galliformes | Odontophoridae Washington
#> 8 Aves | Charadriiformes | Laridae Washington
#> 9 Aves | Charadriiformes | Laridae Washington
#> 10 Aves | Charadriiformes | Laridae Washington
#> # ... with 58 more variables: basisofrecord <chr>, month <chr>,
#> # decimallongitude <chr>, phylum <chr>, references <chr>, year <chr>,
#> # startdayofyear <chr>, taxonrank <chr>, specificepithet <chr>,
#> # bibliographiccitation <chr>, family <chr>, countrycode <chr>,
#> # geodeticdatum <chr>, coordinateuncertaintyinmeters <chr>,
#> # highergeography <chr>, continent <chr>, verbatimlocality <chr>,
#> # day <chr>, kingdom <chr>, collectioncode <chr>,
#> # occurrencestatus <chr>, coordinateprecision <chr>,
#> # institutioncode <chr>, scientificname <chr>, locality <chr>,
#> # class <chr>, vernacularname <chr>, county <chr>,
#> # decimallatitude <chr>, occurrenceid <chr>, language <chr>,
#> # license <chr>, country <chr>, georeferenceverificationstatus <chr>,
#> # modified <chr>, eventdate <chr>, nomenclaturalcode <chr>,
#> # verbatimeventdate <chr>, genus <chr>, order <chr>,
#> # catalognumber <chr>, enddayofyear <chr>, locationremarks <chr>,
#> # infraspecificepithet <chr>, accessrights <chr>, sex <chr>,
#> # institutionid <chr>, georeferenceprotocol <chr>,
#> # georeferenceddate <chr>, georeferencedby <chr>, preparations <chr>,
#> # recordedby <chr>, georeferenceremarks <chr>, dynamicproperties <chr>,
#> # georeferencesources <chr>, othercatalognumbers <chr>,
#> # occurrenceremarks <chr>, lifestage <chr>
Limit the number of records returned (under 1000)
res <- vertsearch("(kansas state OR KSU)", limit = 200)
res$data
#> # A tibble: 200 x 78
#> individualcount georeferenceprotocol
#> <chr> <chr>
#> 1 8 GEOLocate (Rios & Bart, 2010)
#> 2 11 GEOLocate (Rios & Bart, 2010)
#> 3 3 GEOLocate (Rios & Bart, 2010)
#> 4 <NA> <NA>
#> 5 <NA> <NA>
#> 6 <NA> <NA>
#> 7 1 VertNet Georeferencing Guidelines
#> 8 1 VertNet Georeferencing Guidelines
#> 9 1 VertNet Georeferencing Guidelines
#> 10 1 VertNet Georeferencing Guidelines
#> # ... with 190 more rows, and 76 more variables: recordedby <chr>,
#> # bibliographiccitation <chr>, stateprovince <chr>, basisofrecord <chr>,
#> # month <chr>, decimallongitude <chr>, phylum <chr>, references <chr>,
#> # georeferencedby <chr>, year <chr>, taxonrank <chr>,
#> # specificepithet <chr>, family <chr>, countrycode <chr>,
#> # locality <chr>, geodeticdatum <chr>,
#> # coordinateuncertaintyinmeters <chr>, highergeography <chr>,
#> # continent <chr>, day <chr>, kingdom <chr>, georeferenceddate <chr>,
#> # footprintwkt <chr>, institutioncode <chr>, scientificname <chr>,
#> # preparations <chr>, disposition <chr>, class <chr>,
#> # identificationremarks <chr>, county <chr>, decimallatitude <chr>,
#> # occurrenceid <chr>, language <chr>, license <chr>, country <chr>,
#> # georeferenceverificationstatus <chr>, othercatalognumbers <chr>,
#> # infraspecificepithet <chr>, eventdate <chr>, identifiedby <chr>,
#> # nomenclaturalcode <chr>, fieldnumber <chr>, verbatimeventdate <chr>,
#> # genus <chr>, order <chr>, catalognumber <chr>, collectioncode <chr>,
#> # higherclassification <chr>, lifestage <chr>, startdayofyear <chr>,
#> # occurrenceremarks <chr>, verbatimlocality <chr>,
#> # georeferencesources <chr>, verbatimcoordinatesystem <chr>,
#> # institutionid <chr>, modified <chr>, dateidentified <chr>,
#> # enddayofyear <chr>, georeferenceremarks <chr>, accessrights <chr>,
#> # occurrencestatus <chr>, sex <chr>, establishmentmeans <chr>,
#> # identificationverificationstatus <chr>, identificationqualifier <chr>,
#> # organismid <chr>, dynamicproperties <chr>, verbatimcoordinates <chr>,
#> # locationaccordingto <chr>, recordnumber <chr>,
#> # previousidentifications <chr>, samplingprotocol <chr>,
#> # minimumelevationinmeters <chr>, maximumelevationinmeters <chr>,
#> # datasetname <chr>, collectionid <chr>
Pass output of vertsearch() to a map
out <- vertsearch(tax = "(mustela nivalis OR mustela erminea)")
vertmap(out)
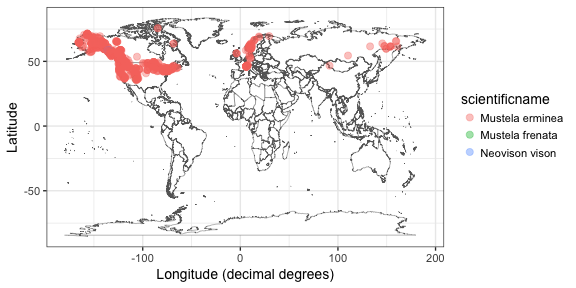
Lots of data
For searchbyterm(), spatialsearch(), and vertsearch(), you can request more than 1000 records. VertNet limits each request to 1000 records, but internally in this package, if you request more than 1000 records, we’ll continue to send requests to get all the records you want. See the VertNet docs for more information on this.
Email dump of data
bigsearch() specifies a termwise search (like searchbyterm()), but requests that all available records be made available for download as a tab-delimited text file.
bigsearch(genus = "ochotona", rfile = "mydata", email = "you@gmail.com")
#> Processing request...
#>
#> Download of records file 'mydata' requested for 'you@gmail.com'
#>
#> Query/URL: "http://api.vertnet-portal.appspot.com/api/download?q=%7B%22q%22:%22genus:ochotona%22,%22n%22:%22mydata%22,%22e%22:%22you@gmail.com%22%7D"
#>
#> Thank you! Download instructions will be sent by email.
Messages
In the previous examples, we’ve suppressed messages for more concise output, but you can set verbose=TRUE to get helpful messages - verbose=TRUE is also the default setting so if you don’t specify that parameter messages will be printed to the console.
res <- searchbyterm(class = "Aves", state = "California", limit = 10, verbose = TRUE)
Citing
To cite rvertnet in publications use:
Scott Chamberlain, Chris Ray and Vijay Barve (2017). rvertnet: Search ‘Vertnet’, a ‘Database’ of Vertebrate Specimen Records. R package version 0.6.2. https://CRAN.R-project.org/package=rvertnet
License and bugs
- License: MIT
- Report bugs at our Github repo for rvertnet
