rplos tutorial
for v0.5.6
The rplos package interacts with the API services of PLoS (Public Library of Science) Journals. You used to need an API key to work with this package - that is no longer needed!
This tutorial will go through three use cases to demonstrate the kinds
of things possible in rplos.
- Search across PLoS papers in various sections of papers
- Search for terms and visualize results as a histogram OR as a plot through time
- Text mining of scientific literature
Installation
install.packages("rplos")
Or development version
if (!require("devtools")) install.packages("devtools")
devtools::install_github("ropensci/rplos")
library("rplos")
Usage
Search across PLoS papers in various sections of papers
searchplos is a general search, and in this case searches for the term
Helianthus and returns the DOI’s of matching papers
searchplos(q= "Helianthus", fl= "id", limit = 5)
#> $meta
#> numFound start maxScore
#> 1 422 0 NA
#>
#> $data
#> id
#> 1 10.1371/journal.pone.0148280
#> 2 10.1371/journal.pone.0111982
#> 3 10.1371/journal.pone.0057533
#> 4 10.1371/journal.pone.0139188
#> 5 10.1371/journal.pone.0045899
Get only full article DOIs
searchplos(q="*:*", fl='id', fq='doc_type:full', start=0, limit=5)
#> $meta
#> numFound start maxScore
#> 1 184983 0 NA
#>
#> $data
#> id
#> 1 10.1371/journal.pone.0107420
#> 2 10.1371/annotation/492fdf80-c999-4947-b569-96af8cb4e9d9
#> 3 10.1371/annotation/7a3d2279-0f96-433c-bb3f-d7fda1759633
#> 4 10.1371/annotation/78d328b9-2c8c-4978-84b1-7e6a0b12ada1
#> 5 10.1371/annotation/7983e1b9-09e4-4123-b1d5-aaaa0121e76a
Get DOIs for only PLoS One articles
searchplos(q="*:*", fl='id', fq='cross_published_journal_key:PLoSONE', start=0, limit=5)
#> $meta
#> numFound start maxScore
#> 1 1344674 0 NA
#>
#> $data
#> id
#> 1 10.1371/journal.pone.0107420
#> 2 10.1371/journal.pone.0107420/title
#> 3 10.1371/journal.pone.0107420/abstract
#> 4 10.1371/journal.pone.0107420/references
#> 5 10.1371/journal.pone.0107420/body
Get DOIs for full article in PLoS One
searchplos(q="*:*", fl='id',
fq=list('cross_published_journal_key:PLoSONE', 'doc_type:full'),
start=0, limit=5)
#> $meta
#> numFound start maxScore
#> 1 157139 0 NA
#>
#> $data
#> id
#> 1 10.1371/journal.pone.0107420
#> 2 10.1371/annotation/492fdf80-c999-4947-b569-96af8cb4e9d9
#> 3 10.1371/annotation/7a3d2279-0f96-433c-bb3f-d7fda1759633
#> 4 10.1371/annotation/78d328b9-2c8c-4978-84b1-7e6a0b12ada1
#> 5 10.1371/annotation/7983e1b9-09e4-4123-b1d5-aaaa0121e76a
Search for many terms
q <- c('ecology','evolution','science')
lapply(q, function(x) searchplos(x, limit=2))
#> [[1]]
#> [[1]]$meta
#> numFound start maxScore
#> 1 32852 0 NA
#>
#> [[1]]$data
#> id
#> 1 10.1371/journal.pone.0059813
#> 2 10.1371/journal.pone.0001248
#>
#>
#> [[2]]
#> [[2]]$meta
#> numFound start maxScore
#> 1 53560 0 NA
#>
#> [[2]]$data
#> id
#> 1 10.1371/annotation/9773af53-a076-4946-a3f1-83914226c10d
#> 2 10.1371/annotation/c55d5089-ba2f-449d-8696-2bc8395978db
#>
#>
#> [[3]]
#> [[3]]$meta
#> numFound start maxScore
#> 1 157210 0 NA
#>
#> [[3]]$data
#> id
#> 1 10.1371/journal.pbio.0020122
#> 2 10.1371/journal.pbio.1001166
Search on specific sections
A suite of functions were created as light wrappers around searchplos as a shorthand to search specific sections of a paper.
plosauthorsearchers in authorsplosabstractsearches in abstractsplostitlesearches in titlesplosfigtabcapssearches in figure and table captionsplossubjectsearches in subject areas
plosauthor searches across authors, and in this case returns the authors of the matching papers. the fl parameter determines what is returned
plosauthor(q = "Eisen", fl = "author", limit = 5)
#> $meta
#> numFound start maxScore
#> 1 833 0 NA
#>
#> $data
#> author
#> 1 Jonathan A Eisen
#> 2 Jonathan A Eisen
#> 3 Jonathan A Eisen
#> 4 Jonathan A Eisen
#> 5 Jonathan A Eisen
plosabstract searches across abstracts, and in this case returns the id and title of the matching papers
plosabstract(q = 'drosophila', fl='id,title', limit = 5)
#> $meta
#> numFound start maxScore
#> 1 2925 0 NA
#>
#> $data
#> id
#> 1 10.1371/journal.pbio.0040198
#> 2 10.1371/journal.pbio.0030246
#> 3 10.1371/journal.pone.0012421
#> 4 10.1371/journal.pbio.0030389
#> 5 10.1371/journal.pone.0002817
#> title
#> 1 All for All
#> 2 School Students as Drosophila Experimenters
#> 3 Host Range and Specificity of the Drosophila C Virus
#> 4 New Environments Set the Stage for Changing Tastes in Mates
#> 5 High-Resolution, In Vivo Magnetic Resonance Imaging of Drosophila at 18.8 Tesla
plostitle searches across titles, and in this case returns the title and journal of the matching papers
plostitle(q='drosophila', fl='title,journal', limit=5)
#> $meta
#> numFound start maxScore
#> 1 1892 0 NA
#>
#> $data
#> journal
#> 1 PLoS Biology
#> 2 PLoS Biology
#> 3 PLoS Genetics
#> 4 PLoS Computational Biology
#> 5 PLoS ONE
#> title
#> 1 Reinforcement of Gametic Isolation in Drosophila
#> 2 Identification of Drosophila MicroRNA Targets
#> 3 Phenotypic Plasticity of the Drosophila Transcriptome
#> 4 Parametric Alignment of Drosophila Genomes
#> 5 A Tripartite Synapse Model in Drosophila
Search for terms and visualize results as a histogram OR as a plot through time
plosword allows you to search for 1 to K words and visualize the results
as a histogram, comparing number of matching papers for each word
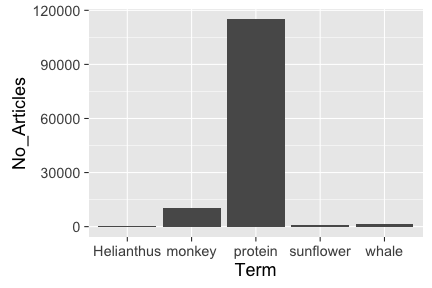
out <- plosword(list("monkey", "Helianthus", "sunflower", "protein", "whale"),
vis = "TRUE")
out$table
#> No_Articles Term
#> 1 10289 monkey
#> 2 422 Helianthus
#> 3 1144 sunflower
#> 4 114997 protein
#> 5 1331 whale
out$plot
You can also pass in curl options, in this case get verbose information on the curl call.
plosword('Helianthus', callopts=list(verbose=TRUE))
#> Number of articles with search term
#> 422
Visualize terms
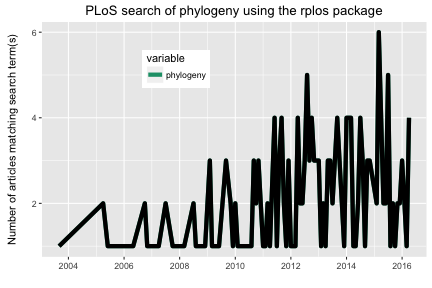
plot_throughtime allows you to search for up to 2 words and visualize the results as a line plot through time, comparing number of articles matching through time. Visualize with the ggplot2 package, only up to two terms for now.
library("ggplot2")
plot_throughtime(terms = "phylogeny", limit = 200) + geom_line(size=2, color='black')
Faceted search
In addition to searchplos() and related searching functions, there are a few slightly different ways to search: faceting and highlighted searches. Faceting allows you to ask, e.g., how many articles are published in each of the PLOS journals. Highlighting allows you to ask, e.g., highlight terms that I search for in the text results given back, which can make downstream processing easier, and help visualize search results (see highbrow() below).
Facet by journal
facetplos(q='*:*', facet.field='journal')
#> $facet_queries
#> NULL
#>
#> $facet_fields
#> $facet_fields$journal
#> X1 X2
#> 1 plos one 1285831
#> 2 plos genetics 50706
#> 3 plos pathogens 44134
#> 4 plos computational biology 37646
#> 5 plos neglected tropical diseases 35979
#> 6 plos biology 29392
#> 7 plos medicine 20335
#> 8 plos clinical trials 521
#> 9 plos medicin 9
#>
#>
#> $facet_dates
#> NULL
#>
#> $facet_ranges
#> NULL
Using facet.query to get counts
facetplos(q='*:*', facet.field='journal', facet.query='cell,bird')
#> $facet_queries
#> term value
#> 1 cell,bird 24
#>
#> $facet_fields
#> $facet_fields$journal
#> X1 X2
#> 1 plos one 1285831
#> 2 plos genetics 50706
#> 3 plos pathogens 44134
#> 4 plos computational biology 37646
#> 5 plos neglected tropical diseases 35979
#> 6 plos biology 29392
#> 7 plos medicine 20335
#> 8 plos clinical trials 521
#> 9 plos medicin 9
#>
#>
#> $facet_dates
#> NULL
#>
#> $facet_ranges
#> NULL
Date faceting
facetplos(q='*:*', url=url, facet.date='publication_date',
facet.date.start='NOW/DAY-5DAYS', facet.date.end='NOW', facet.date.gap='+1DAY')
#> $facet_queries
#> NULL
#>
#> $facet_fields
#> NULL
#>
#> $facet_dates
#> $facet_dates$publication_date
#> date value
#> 1 2016-04-27T00:00:00Z 2247
#> 2 2016-04-28T00:00:00Z 2187
#> 3 2016-04-29T00:00:00Z 848
#> 4 2016-04-30T00:00:00Z 0
#> 5 2016-05-01T00:00:00Z 0
#> 6 2016-05-02T00:00:00Z 0
#>
#>
#> $facet_ranges
#> NULL
Highlighted search
Search for the term alcohol in the abstracts of articles, return only 10 results
highplos(q='alcohol', hl.fl = 'abstract', rows=2)
#> $`10.1371/journal.pmed.0040151`
#> $`10.1371/journal.pmed.0040151`$abstract
#> [1] "Background: <em>Alcohol</em> consumption causes an estimated 4% of the global disease burden, prompting"
#>
#>
#> $`10.1371/journal.pone.0027752`
#> $`10.1371/journal.pone.0027752`$abstract
#> [1] "Background: The negative influences of <em>alcohol</em> on TB management with regard to delays in seeking"
Search for the term alcohol in the abstracts of articles, and return fragment size of 20 characters, return only 5 results
highplos(q='alcohol', hl.fl='abstract', hl.fragsize=20, rows=2)
#> $`10.1371/journal.pmed.0040151`
#> $`10.1371/journal.pmed.0040151`$abstract
#> [1] "Background: <em>Alcohol</em>"
#>
#>
#> $`10.1371/journal.pone.0027752`
#> $`10.1371/journal.pone.0027752`$abstract
#> [1] " of <em>alcohol</em> on TB management"
Search for the term experiment across all sections of an article, return id (DOI) and title fl only, search in full articles only (via fq='doc_type:full'), and return only 10 results
highplos(q='everything:"experiment"', fl='id,title', fq='doc_type:full',
rows=2)
#> $`10.1371/journal.pone.0154334`
#> $`10.1371/journal.pone.0154334`$everything
#> [1] " and designed the <em>experiments</em>: RJ CM AOC. Performed the <em>experiments</em>: RJ AOC. Analyzed the data: RJ. Contributed"
#>
#>
#> $`10.1371/journal.pone.0039681`
#> $`10.1371/journal.pone.0039681`$everything
#> [1] " Selection of Transcriptomics <em>Experiments</em> Improves Guilt-by-Association Analyses Transcriptomics <em>Experiment</em>"
Visualize highligted searches
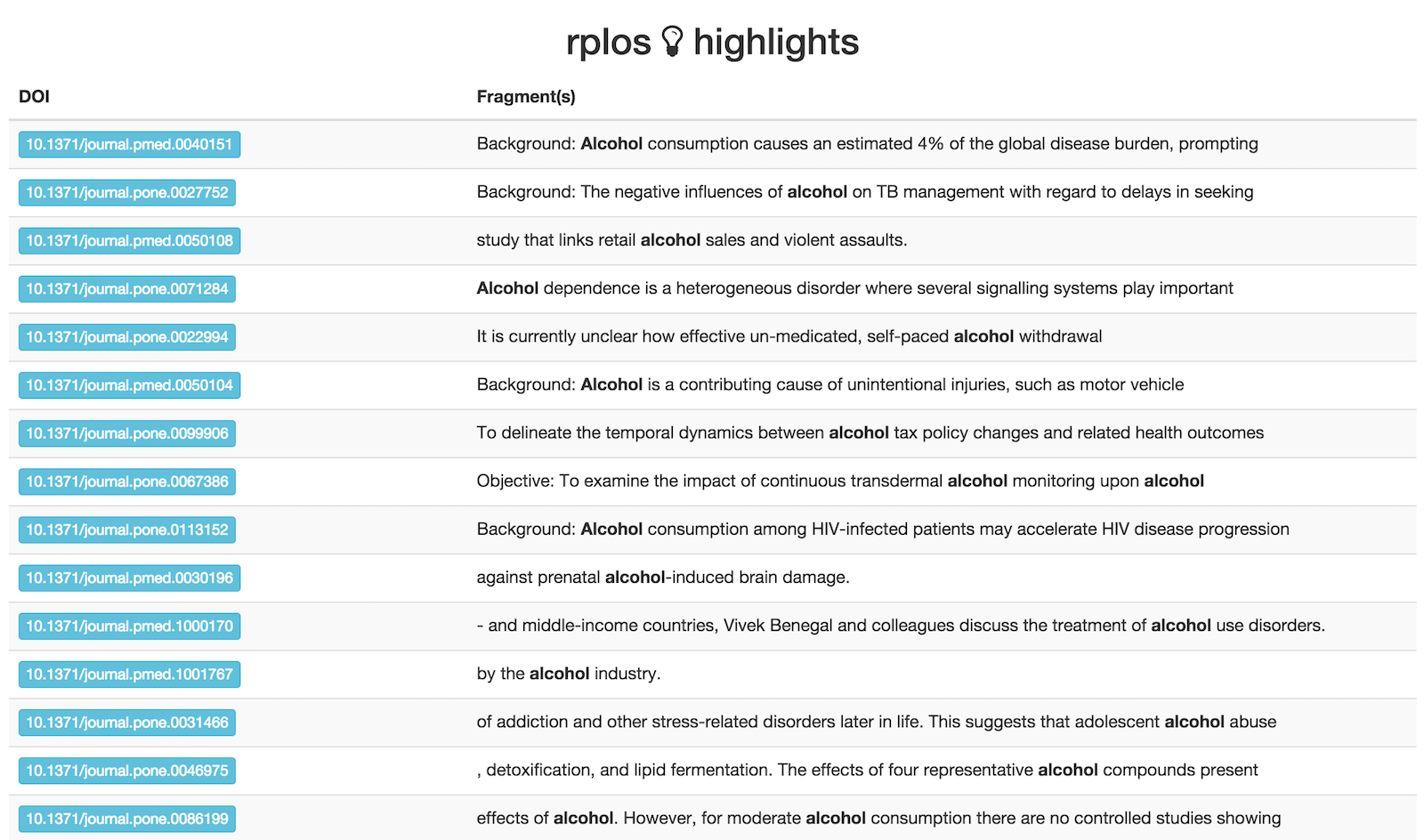
Browse highlighted fragments in your default browser
This first examle, we only looko at 10 results
out <- highplos(q='alcohol', hl.fl = 'abstract', rows=10)
highbrow(out)

But it works quickly with lots of results too
out <- highplos(q='alcohol', hl.fl = 'abstract', rows=1200)
highbrow(out)
Citing
To cite rplos in publications use:
Scott Chamberlain, Carl Boettiger and Karthik Ram (2016). rplos: Interface to PLOS Journals search API. R package version 0.5.6 https://github.com/ropensci/rplos
License and bugs
- License: MIT
- Report bugs at our Github repo for rplos
