rgbif tutorial
for v0.9.9
Seach and retrieve data from the Global Biodiverity Information Facilty (GBIF)
rgbif is an R package to search and retrieve data from the Global Biodiverity Information Facilty (GBIF). rgbif wraps R code around the GBIF API to allow you to talk to GBIF from R.
Installation
Install from CRAN
install.packages("rgbif")
Or install the development version from GitHub
devtools::install_github("ropensci/rgbif")
Load rgbif
library("rgbif")
Number of occurrences
Search by type of record, all observational in this case
occ_count(basisOfRecord='OBSERVATION')
#> [1] 30364566
Records for Puma concolor with lat/long data (georeferened) only. Note that hasCoordinate in occ_search() is the same as georeferenced in occ_count().
occ_count(taxonKey=2435099, georeferenced=TRUE)
#> [1] 3747
All georeferenced records in GBIF
occ_count(georeferenced=TRUE)
#> [1] 772524989
Records from Denmark
denmark_code <- isocodes[grep("Denmark", isocodes$name), "code"]
occ_count(country=denmark_code)
#> [1] 11372130
Number of records in a particular dataset
occ_count(datasetKey='9e7ea106-0bf8-4087-bb61-dfe4f29e0f17')
#> [1] 4591
All records from 2012
occ_count(year=2012)
#> [1] 44688340
Records for a particular dataset, and only for preserved specimens
occ_count(datasetKey='e707e6da-e143-445d-b41d-529c4a777e8b', basisOfRecord='OBSERVATION')
#> [1] 0
Search for taxon names
Get possible values to be used in taxonomic rank arguments in functions
taxrank()
#> [1] "kingdom" "phylum" "class" "order"
#> [5] "family" "genus" "species" "infraspecific"
name_lookup() does full text search of name usages covering the scientific and vernacular name, the species description, distribution and the entire classification across all name usages of all or some checklists. Results are ordered by relevance as this search usually returns a lot of results.
By default name_lookup() returns five slots of information: meta, data, facets, hierarchies, and names. hierarchies and names elements are named by their matching GBIF key in the data.frame in the data slot.
out <- name_lookup(query='mammalia')
names(out)
#> [1] "meta" "data" "facets" "hierarchies" "names"
out$meta
#> # A tibble: 1 x 4
#> offset limit endOfRecords count
#> <int> <int> <lgl> <int>
#> 1 0 100 FALSE 1329
head(out$data)
#> # A tibble: 6 x 28
#> key scientificName datasetKey nubKey
#> <int> <chr> <chr> <int>
#> 1 114079693 Mammalia bd0a2b6d-69d1-4650-8bb1-829c8f92035f 359
#> 2 123227024 Mammalia 90d9e8a6-0ce1-472d-b682-3451095dbc5a 359
#> 3 115498126 Mammalia fe51a93b-a7f8-48ec-99c8-7c347716e8d3 359
#> 4 127776750 Mammalia 5b3a293b-04e5-4ad7-992f-cb016fcf166a 359
#> 5 100375341 Mammalia 16c3f9cb-4b19-4553-ac8e-ebb90003aa02 359
#> 6 133364782 Mammalia 7ddf754f-d193-4cc9-b351-99906754a03b 359
#> # ... with 24 more variables: parentKey <int>, parent <chr>,
#> # canonicalName <chr>, nameType <chr>, taxonomicStatus <chr>,
#> # origin <chr>, numDescendants <int>, numOccurrences <int>,
#> # taxonID <chr>, habitats <chr>, nomenclaturalStatus <lgl>,
#> # threatStatuses <lgl>, synonym <lgl>, authorship <chr>, kingdom <chr>,
#> # phylum <chr>, kingdomKey <int>, phylumKey <int>, classKey <int>,
#> # rank <chr>, class <chr>, extinct <lgl>, constituentKey <chr>,
#> # publishedIn <chr>
out$facets
#> NULL
out$hierarchies[1:2]
#> $`114079693`
#> rankkey name
#> 1 114079410 Animalia
#>
#> $`123227024`
#> rankkey name
#> 1 123221530 Gnathostomata
out$names[2]
#> $`359`
#> vernacularName language
#> 1 zoogdieren nld
#> 2 Mammals eng
#> 3 mammal eng
#> 4 mammals
#> 5 däggdjur swe
#> 6 mammals eng
#> 7 mammifères fra
#> 8 pattedyr nno
#> 9 pattedyr nob
#> 10 sogdyr nno
#> 11 spendyr nno
#> 12 zoogdieren nld
#> 13 Ссавці ukr
#> 14 哺乳 jpn
#> 15 mammals eng
#> 16 mammifères fra
#> 17 mamÃfero por
Search for a genus
head(name_lookup(query='Cnaemidophorus', rank="genus", return="data"))
#> # A tibble: 6 x 34
#> key scientificName datasetKey nubKey
#> <int> <chr> <chr> <int>
#> 1 133063907 Cnaemidophorus 4cec8fef-f129-4966-89b7-4f8439aba058 1858636
#> 2 134031474 Cnaemidophorus 23905003-5ee5-4326-b1bb-3a2fd6250df3 1858636
#> 3 133419671 Cnaemidophorus 7ddf754f-d193-4cc9-b351-99906754a03b 1858636
#> 4 135314362 Cnaemidophorus 16c3f9cb-4b19-4553-ac8e-ebb90003aa02 1858636
#> 5 135332978 Cnaemidophorus cbb6498e-8927-405a-916b-576d00a6289b 1858636
#> 6 135447727 Cnaemidophorus d16563e0-e718-45a9-a20f-3e9fc20613da 1858636
#> # ... with 30 more variables: parentKey <int>, parent <chr>, phylum <chr>,
#> # order <chr>, family <chr>, genus <chr>, phylumKey <int>,
#> # classKey <int>, orderKey <int>, familyKey <int>, genusKey <int>,
#> # canonicalName <chr>, taxonomicStatus <chr>, rank <chr>, origin <chr>,
#> # numDescendants <int>, numOccurrences <int>, habitats <lgl>,
#> # nomenclaturalStatus <lgl>, threatStatuses <lgl>, synonym <lgl>,
#> # class <chr>, kingdom <chr>, kingdomKey <int>, nameType <chr>,
#> # taxonID <chr>, authorship <chr>, publishedIn <chr>, extinct <lgl>,
#> # constituentKey <chr>
Search for the class mammalia
head(name_lookup(query='mammalia', return = 'data'))
#> # A tibble: 6 x 28
#> key scientificName datasetKey nubKey
#> <int> <chr> <chr> <int>
#> 1 114079693 Mammalia bd0a2b6d-69d1-4650-8bb1-829c8f92035f 359
#> 2 123227024 Mammalia 90d9e8a6-0ce1-472d-b682-3451095dbc5a 359
#> 3 115498126 Mammalia fe51a93b-a7f8-48ec-99c8-7c347716e8d3 359
#> 4 127776750 Mammalia 5b3a293b-04e5-4ad7-992f-cb016fcf166a 359
#> 5 100375341 Mammalia 16c3f9cb-4b19-4553-ac8e-ebb90003aa02 359
#> 6 133364782 Mammalia 7ddf754f-d193-4cc9-b351-99906754a03b 359
#> # ... with 24 more variables: parentKey <int>, parent <chr>,
#> # canonicalName <chr>, nameType <chr>, taxonomicStatus <chr>,
#> # origin <chr>, numDescendants <int>, numOccurrences <int>,
#> # taxonID <chr>, habitats <chr>, nomenclaturalStatus <lgl>,
#> # threatStatuses <lgl>, synonym <lgl>, authorship <chr>, kingdom <chr>,
#> # phylum <chr>, kingdomKey <int>, phylumKey <int>, classKey <int>,
#> # rank <chr>, class <chr>, extinct <lgl>, constituentKey <chr>,
#> # publishedIn <chr>
Look up the species Helianthus annuus
head(name_lookup(query = 'Helianthus annuus', rank="species", return = 'data'))
#> # A tibble: 6 x 42
#> key scientificName datasetKey
#> <int> <chr> <chr>
#> 1 3119195 Helianthus annuus d7dddbf4-2cf0-4f39-9b2a-bb099caae36c
#> 2 134854463 Helianthus annuus f82a4f7f-6f84-4b58-82e6-6b41ec9a1f49
#> 3 114910965 Helianthus annuus ee2aac07-de9a-47a2-b828-37430d537633
#> 4 134843454 Helianthus annuus 29d2d5a6-db22-4abd-b784-9ab2f9757c3c
#> 5 127670355 Helianthus annuus 41c06f1a-23da-4445-b859-ec3a8a03b0e2
#> 6 103340289 Helianthus annuus fab88965-e69d-4491-a04d-e3198b626e52
#> # ... with 39 more variables: constituentKey <chr>, nubKey <int>,
#> # parentKey <int>, parent <chr>, kingdom <chr>, phylum <chr>,
#> # order <chr>, family <chr>, genus <chr>, species <chr>,
#> # kingdomKey <int>, phylumKey <int>, classKey <int>, orderKey <int>,
#> # familyKey <int>, genusKey <int>, speciesKey <int>,
#> # canonicalName <chr>, authorship <chr>, publishedIn <chr>,
#> # nameType <chr>, taxonomicStatus <chr>, rank <chr>, origin <chr>,
#> # numDescendants <int>, numOccurrences <int>, extinct <lgl>,
#> # habitats <chr>, nomenclaturalStatus <chr>, threatStatuses <lgl>,
#> # synonym <lgl>, class <chr>, taxonID <chr>, acceptedKey <int>,
#> # accepted <chr>, accordingTo <chr>, nameKey <int>, basionymKey <int>,
#> # basionym <chr>
The function name_usage() works with lots of different name endpoints in GBIF, listed at http://www.gbif.org/developer/species#nameUsages.
library("plyr")
out <- name_usage(key=3119195, language="FRENCH", data='vernacularNames')
head(out$data)
#> # A tibble: 6 x 6
#> taxonKey vernacularName language
#> <int> <chr> <chr>
#> 1 3119195 common sunflower eng
#> 2 3119195 tournesol fra
#> 3 3119195 garden sunflower eng
#> 4 3119195 grand soleil fra
#> 5 3119195 hélianthe annuel fra
#> 6 3119195 soleil fra
#> # ... with 3 more variables: source <chr>, sourceTaxonKey <int>,
#> # preferred <lgl>
The function name_backbone() is used to search against the GBIF backbone taxonomy
name_backbone(name='Helianthus', rank='genus', kingdom='plants')
#> $usageKey
#> [1] 3119134
#>
#> $scientificName
#> [1] "Helianthus L."
#>
#> $canonicalName
#> [1] "Helianthus"
#>
#> $rank
#> [1] "GENUS"
#>
#> $status
#> [1] "ACCEPTED"
#>
#> $confidence
#> [1] 97
#>
#> $matchType
#> [1] "EXACT"
#>
#> $kingdom
#> [1] "Plantae"
#>
#> $phylum
#> [1] "Tracheophyta"
#>
#> $order
#> [1] "Asterales"
#>
#> $family
#> [1] "Asteraceae"
#>
#> $genus
#> [1] "Helianthus"
#>
#> $kingdomKey
#> [1] 6
#>
#> $phylumKey
#> [1] 7707728
#>
#> $classKey
#> [1] 220
#>
#> $orderKey
#> [1] 414
#>
#> $familyKey
#> [1] 3065
#>
#> $genusKey
#> [1] 3119134
#>
#> $synonym
#> [1] FALSE
#>
#> $class
#> [1] "Magnoliopsida"
The function name_suggest() is optimized for speed, and gives back suggested names based on query parameters.
head( name_suggest(q='Puma concolor') )
#> # A tibble: 6 x 3
#> key canonicalName rank
#> <int> <chr> <chr>
#> 1 2435099 Puma concolor SPECIES
#> 2 8951716 Puma concolor borbensis SUBSPECIES
#> 3 6164618 Puma concolor browni SUBSPECIES
#> 4 8860878 Puma concolor capricornensis SUBSPECIES
#> 5 8916934 Puma concolor bangsi SUBSPECIES
#> 6 6164599 Puma concolor azteca SUBSPECIES
Single occurrence records
Get data for a single occurrence. Note that data is returned as a list, with slots for metadata and data, or as a hierarchy, or just data.
Just data
occ_get(key=240713150, return='data')
#> name key decimalLatitude decimalLongitude issues
#> 1 Pelosina 240713150 -77.5667 163.583 bri,cdround,gass84
Just taxonomic hierarchy
occ_get(key=240713150, return='hier')
#> name key rank
#> 1 Chromista 4 kingdom
#> 2 Foraminifera 8376456 phylum
#> 3 Monothalamea 7882876 class
#> 4 Astrorhizida 8142878 order
#> 5 Astrorhizidae 7747923 family
#> 6 Pelosina 7822114 genus
All data, or leave return parameter blank
occ_get(key=240713150, return='all')
#> $hierarchy
#> name key rank
#> 1 Chromista 4 kingdom
#> 2 Foraminifera 8376456 phylum
#> 3 Monothalamea 7882876 class
#> 4 Astrorhizida 8142878 order
#> 5 Astrorhizidae 7747923 family
#> 6 Pelosina 7822114 genus
#>
#> $media
#> list()
#>
#> $data
#> name key decimalLatitude decimalLongitude issues
#> 1 Pelosina 240713150 -77.5667 163.583 bri,cdround,gass84
Get many occurrences. occ_get is vectorized
occ_get(key=c(101010, 240713150, 855998194), return='data')
#> name key decimalLatitude decimalLongitude
#> 1 Platydoras armatulus 101010 NA NA
#> 2 Pelosina 240713150 -77.56670 163.58299
#> 3 Sciurus vulgaris 855998194 58.40677 12.04386
#> issues
#> 1
#> 2 bri,cdround,gass84
#> 3 cdround,gass84,rdatm
Search for occurrences
By default occ_search() returns a dplyr like output summary in which the data printed expands based on how much data is returned, and the size of your window. You can search by scientific name:
occ_search(scientificName = "Ursus americanus", limit = 20)
#> Records found [9455]
#> Records returned [20]
#> No. unique hierarchies [1]
#> No. media records [16]
#> No. facets [0]
#> Args [limit=20, offset=0, scientificName=Ursus americanus, fields=all]
#> # A tibble: 20 x 66
#> name key decimalLatitude decimalLongitude
#> <chr> <int> <dbl> <dbl>
#> 1 Ursus americanus 1671722060 33.47152 -91.64752
#> 2 Ursus americanus 1453325042 37.36325 -80.52914
#> 3 Ursus americanus 1453341157 35.44519 -83.75077
#> 4 Ursus americanus 1453341156 35.43836 -83.66423
#> 5 Ursus americanus 1453427952 35.61469 -82.47723
#> 6 Ursus americanus 1453476835 29.24034 -103.30502
#> 7 Ursus americanus 1453456359 25.31110 -100.96992
#> 8 Ursus americanus 1453414927 47.90953 -91.95893
#> 9 Ursus americanus 1453456338 25.30959 -100.96966
#> 10 Ursus americanus 1453445710 35.59506 -82.55149
#> 11 Ursus americanus 1457591001 38.93946 -119.94679
#> 12 Ursus americanus 1640034659 39.20098 -123.16410
#> 13 Ursus americanus 1500285318 45.51560 -69.35556
#> 14 Ursus americanus 1455592330 46.34195 -83.98219
#> 15 Ursus americanus 1571069736 25.25062 -100.94652
#> 16 Ursus americanus 1500319247 32.89649 -109.48065
#> 17 Ursus americanus 1453476285 35.94219 -76.57357
#> 18 Ursus americanus 1457595069 46.59933 -84.46328
#> 19 Ursus americanus 1500222058 25.34884 -100.91091
#> 20 Ursus americanus 1500223333 45.38687 -76.10825
#> # ... with 62 more variables: issues <chr>, datasetKey <chr>,
#> # publishingOrgKey <chr>, publishingCountry <chr>, protocol <chr>,
#> # lastCrawled <chr>, lastParsed <chr>, crawlId <int>, extensions <chr>,
#> # basisOfRecord <chr>, taxonKey <int>, kingdomKey <int>,
#> # phylumKey <int>, classKey <int>, orderKey <int>, familyKey <int>,
#> # genusKey <int>, speciesKey <int>, scientificName <chr>, kingdom <chr>,
#> # phylum <chr>, order <chr>, family <chr>, genus <chr>, species <chr>,
#> # genericName <chr>, specificEpithet <chr>, taxonRank <chr>,
#> # dateIdentified <chr>, coordinateUncertaintyInMeters <dbl>, year <int>,
#> # month <int>, day <int>, eventDate <chr>, modified <chr>,
#> # lastInterpreted <chr>, references <chr>, license <chr>,
#> # identifiers <chr>, facts <chr>, relations <chr>, geodeticDatum <chr>,
#> # class <chr>, countryCode <chr>, country <chr>, rightsHolder <chr>,
#> # identifier <chr>, verbatimEventDate <chr>, datasetName <chr>,
#> # verbatimLocality <chr>, gbifID <chr>, collectionCode <chr>,
#> # occurrenceID <chr>, taxonID <chr>, catalogNumber <chr>,
#> # recordedBy <chr>, http...unknown.org.occurrenceDetails <chr>,
#> # institutionCode <chr>, rights <chr>, identificationID <chr>,
#> # eventTime <chr>, occurrenceRemarks <chr>
Or to be more precise, you can search for names first, make sure you have the right name, then pass the GBIF key to the occ_search() function:
key <- name_suggest(q='Helianthus annuus', rank='species')$key[1]
occ_search(taxonKey=key, limit=20)
#> Records found [18310]
#> Records returned [20]
#> No. unique hierarchies [1]
#> No. media records [1]
#> No. facets [0]
#> Args [limit=20, offset=0, taxonKey=9206251, fields=all]
#> # A tibble: 20 x 88
#> name key decimalLatitude decimalLongitude
#> <chr> <int> <dbl> <dbl>
#> 1 Helianthus annuus 1433793045 59.66859 16.54257
#> 2 Helianthus annuus 1434024463 63.71622 20.31247
#> 3 Helianthus annuus 1563876655 NA NA
#> 4 Helianthus annuus 1436147509 59.85465 17.79089
#> 5 Helianthus annuus 1436223234 59.85509 17.78900
#> 6 Helianthus annuus 1450388036 56.60630 16.64841
#> 7 Helianthus annuus 1499896133 58.76637 16.24997
#> 8 Helianthus annuus 1499929475 59.85530 17.79055
#> 9 Helianthus annuus 1669229145 59.85530 17.79055
#> 10 Helianthus annuus 1669043510 59.74332 17.78161
#> 11 Helianthus annuus 1669900943 57.73119 16.13173
#> 12 Helianthus annuus 1669884935 56.64750 12.85256
#> 13 Helianthus annuus 1669797382 55.72320 13.18387
#> 14 Helianthus annuus 1669882522 58.53514 16.50554
#> 15 Helianthus annuus 1670276641 57.30363 11.90345
#> 16 Helianthus annuus 1669981909 57.72138 11.94415
#> 17 Helianthus annuus 1670011022 59.85046 17.68560
#> 18 Helianthus annuus 1670291603 59.15281 18.17448
#> 19 Helianthus annuus 1670396073 59.70498 16.51416
#> 20 Helianthus annuus 1670269738 59.70552 16.20511
#> # ... with 84 more variables: issues <chr>, datasetKey <chr>,
#> # publishingOrgKey <chr>, publishingCountry <chr>, protocol <chr>,
#> # lastCrawled <chr>, lastParsed <chr>, crawlId <int>, extensions <chr>,
#> # basisOfRecord <chr>, individualCount <int>, taxonKey <int>,
#> # kingdomKey <int>, phylumKey <int>, classKey <int>, orderKey <int>,
#> # familyKey <int>, genusKey <int>, speciesKey <int>,
#> # scientificName <chr>, kingdom <chr>, phylum <chr>, order <chr>,
#> # family <chr>, genus <chr>, species <chr>, genericName <chr>,
#> # specificEpithet <chr>, taxonRank <chr>,
#> # coordinateUncertaintyInMeters <dbl>, continent <chr>, year <int>,
#> # month <int>, day <int>, eventDate <chr>, modified <chr>,
#> # lastInterpreted <chr>, license <chr>, identifiers <chr>, facts <chr>,
#> # relations <chr>, geodeticDatum <chr>, class <chr>, countryCode <chr>,
#> # country <chr>, rightsHolder <chr>, county <chr>, municipality <chr>,
#> # identificationVerificationStatus <chr>, gbifID <chr>, language <chr>,
#> # type <chr>, taxonID <chr>, occurrenceStatus <chr>,
#> # catalogNumber <chr>, vernacularName <chr>, institutionCode <chr>,
#> # taxonConceptID <chr>, eventTime <chr>, identifier <chr>,
#> # informationWithheld <chr>, endDayOfYear <chr>, locality <chr>,
#> # collectionCode <chr>, occurrenceID <chr>, recordedBy <chr>,
#> # startDayOfYear <chr>, datasetID <chr>, bibliographicCitation <chr>,
#> # accessRights <chr>, higherClassification <chr>, dateIdentified <chr>,
#> # elevation <dbl>, stateProvince <chr>, references <chr>,
#> # recordNumber <chr>, habitat <chr>, verbatimEventDate <chr>,
#> # associatedTaxa <chr>, verbatimLocality <chr>, verbatimElevation <chr>,
#> # identifiedBy <chr>, identificationID <chr>, occurrenceRemarks <chr>
Like many functions in rgbif, you can choose what to return with the return parameter, here, just returning the metadata:
occ_search(taxonKey=key, return='meta')
#> # A tibble: 1 x 4
#> offset limit endOfRecords count
#> * <int> <int> <lgl> <int>
#> 1 300 200 FALSE 18310
You can choose what fields to return. This isn’t passed on to the API query to GBIF as they don’t allow that, but we filter out the columns before we give the data back to you.
occ_search(scientificName = "Ursus americanus", fields=c('name','basisOfRecord','protocol'), limit = 20)
#> Records found [9455]
#> Records returned [20]
#> No. unique hierarchies [1]
#> No. media records [16]
#> No. facets [0]
#> Args [limit=20, offset=0, scientificName=Ursus americanus,
#> fields=name,basisOfRecord,protocol]
#> # A tibble: 20 x 3
#> name protocol basisOfRecord
#> <chr> <chr> <chr>
#> 1 Ursus americanus DWC_ARCHIVE HUMAN_OBSERVATION
#> 2 Ursus americanus DWC_ARCHIVE HUMAN_OBSERVATION
#> 3 Ursus americanus DWC_ARCHIVE HUMAN_OBSERVATION
#> 4 Ursus americanus DWC_ARCHIVE HUMAN_OBSERVATION
#> 5 Ursus americanus DWC_ARCHIVE HUMAN_OBSERVATION
#> 6 Ursus americanus DWC_ARCHIVE HUMAN_OBSERVATION
#> 7 Ursus americanus DWC_ARCHIVE HUMAN_OBSERVATION
#> 8 Ursus americanus DWC_ARCHIVE HUMAN_OBSERVATION
#> 9 Ursus americanus DWC_ARCHIVE HUMAN_OBSERVATION
#> 10 Ursus americanus DWC_ARCHIVE HUMAN_OBSERVATION
#> 11 Ursus americanus DWC_ARCHIVE HUMAN_OBSERVATION
#> 12 Ursus americanus DWC_ARCHIVE HUMAN_OBSERVATION
#> 13 Ursus americanus DWC_ARCHIVE HUMAN_OBSERVATION
#> 14 Ursus americanus DWC_ARCHIVE HUMAN_OBSERVATION
#> 15 Ursus americanus DWC_ARCHIVE HUMAN_OBSERVATION
#> 16 Ursus americanus DWC_ARCHIVE HUMAN_OBSERVATION
#> 17 Ursus americanus DWC_ARCHIVE HUMAN_OBSERVATION
#> 18 Ursus americanus DWC_ARCHIVE HUMAN_OBSERVATION
#> 19 Ursus americanus DWC_ARCHIVE HUMAN_OBSERVATION
#> 20 Ursus americanus DWC_ARCHIVE HUMAN_OBSERVATION
Most parameters are vectorized, so you can pass in more than one value:
splist <- c('Cyanocitta stelleri', 'Junco hyemalis', 'Aix sponsa')
keys <- sapply(splist, function(x) name_suggest(x)$key[1], USE.NAMES=FALSE)
occ_search(taxonKey=keys, limit=5)
#> Occ. found [2482598 (578358), 2492010 (3062104), 2498387 (978759)]
#> Occ. returned [2482598 (5), 2492010 (5), 2498387 (5)]
#> No. unique hierarchies [2482598 (1), 2492010 (1), 2498387 (1)]
#> No. media records [2482598 (5), 2492010 (5), 2498387 (5)]
#> No. facets [2482598 (0), 2492010 (0), 2498387 (0)]
#> Args [limit=5, offset=0, taxonKey=2482598,2492010,2498387, fields=all]
#> 3 requests; First 10 rows of data from 2482598
#>
#> # A tibble: 5 x 67
#> name key decimalLatitude decimalLongitude
#> <chr> <int> <dbl> <dbl>
#> 1 Cyanocitta stelleri 1453342305 46.14702 -123.6545
#> 2 Cyanocitta stelleri 1453373156 23.67272 -105.4548
#> 3 Cyanocitta stelleri 1453355963 37.76899 -122.4758
#> 4 Cyanocitta stelleri 1453325920 38.48213 -122.8734
#> 5 Cyanocitta stelleri 1453338402 45.50661 -122.7135
#> # ... with 63 more variables: issues <chr>, datasetKey <chr>,
#> # publishingOrgKey <chr>, publishingCountry <chr>, protocol <chr>,
#> # lastCrawled <chr>, lastParsed <chr>, crawlId <int>, extensions <chr>,
#> # basisOfRecord <chr>, taxonKey <int>, kingdomKey <int>,
#> # phylumKey <int>, classKey <int>, orderKey <int>, familyKey <int>,
#> # genusKey <int>, speciesKey <int>, scientificName <chr>, kingdom <chr>,
#> # phylum <chr>, order <chr>, family <chr>, genus <chr>, species <chr>,
#> # genericName <chr>, specificEpithet <chr>, taxonRank <chr>,
#> # dateIdentified <chr>, coordinateUncertaintyInMeters <dbl>, year <int>,
#> # month <int>, day <int>, eventDate <chr>, modified <chr>,
#> # lastInterpreted <chr>, references <chr>, license <chr>,
#> # identifiers <chr>, facts <chr>, relations <chr>, geodeticDatum <chr>,
#> # class <chr>, countryCode <chr>, country <chr>, rightsHolder <chr>,
#> # identifier <chr>, verbatimEventDate <chr>, datasetName <chr>,
#> # verbatimLocality <chr>, gbifID <chr>, collectionCode <chr>,
#> # occurrenceID <chr>, taxonID <chr>, catalogNumber <chr>,
#> # recordedBy <chr>, http...unknown.org.occurrenceDetails <chr>,
#> # institutionCode <chr>, rights <chr>, eventTime <chr>,
#> # identificationID <chr>, occurrenceRemarks <chr>,
#> # informationWithheld <chr>
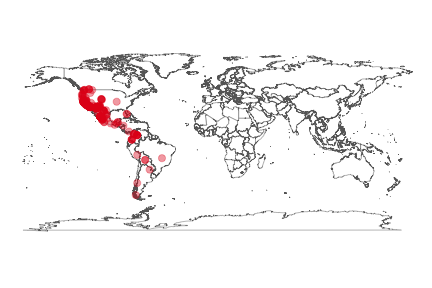
Maps
Static map using the ggplot2 package. Make a map of Puma concolor occurrences.
key <- name_backbone(name='Puma concolor')$speciesKey
dat <- occ_search(taxonKey=key, return='data', limit=300)
gbifmap(dat)
Citing
To cite rgbif in publications use:
Scott Chamberlain, Carl Boettiger, Karthik Ram, Vijay Barve and Dan Mcglinn (2016). rgbif: Interface to the Global Biodiversity Information Facility API. R package version 0.9.3 https://github.com/ropensci/rgbif
License and bugs
- License: MIT
- Report bugs at our Github repo for rgbif
