brranching tutorial
for v0.2.0
R client to fetch phylogenies from many places. Right now we only fetch phylogenies from Phylomatic, but we’d like to add more sources down the road.
Installation
Install and load brranching into the R session. Stable version from CRAN
install.packages("brranching")
Or development version from Github:
install.packages("devtools")
devtools::install_github("ropensci/brranching")
library('brranching')
Usage
Phylomatic
Use the web service
taxa <- c("Poa annua", "Phlox diffusa", "Helianthus annuus")
tree <- phylomatic(taxa = taxa, get = 'POST')
plot(tree, no.margin = TRUE)
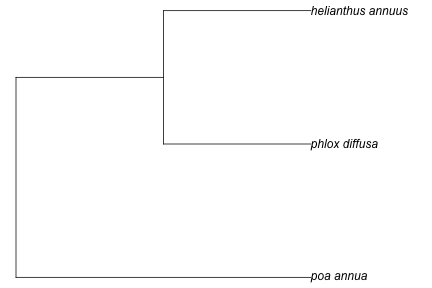
You can pass in up to about 5000 names. We can use taxize to get a random set of plant species names.
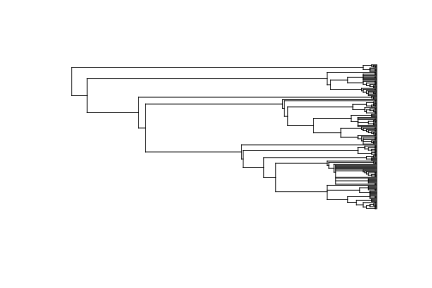
library("taxize")
spp <- names_list("species", 200)
out <- phylomatic(taxa = spp, get = "POST")
plot(out, show.tip.label = FALSE)
Use Phylomatic locally
Phylomatic is written in Awk, and the code can be downloaded to run locally on your own machine.
This approach is for the more adventurous user. The benefit of using Phylomatic locally is
that you can run larger set of names through - when using the web service it has a maximum number
of taxa it will take in as the maintainer does not want any one person taking up a large
amount of server capacity.
First, download the code by doing git clone https://github.com/camwebb/phylomatic-ws, which
will result in a folder phylomatic-ws (or download a zip file, and uncompress it). Then
give the path to that folder in the path parameter in the phylomatic_local() function as
show below.
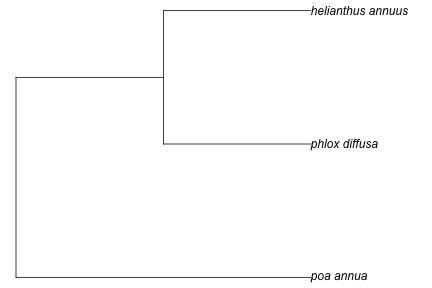
taxa <- c("Poa annua", "Phlox diffusa", "Helianthus annuus")
(tree <- phylomatic_local(taxa, path = "~/github/play/phylomatic-ws"))
#>
#> Phylogenetic tree with 3 tips and 2 internal nodes.
#>
#> Tip labels:
#> [1] "poa_annua" "phlox_diffusa" "helianthus_annuus"
#> Node labels:
#> [1] "poales_to_asterales" "ericales_to_asterales"
#>
#> Rooted; no branch lengths.
Note how the path ~/github/play/phylomatic-ws is specific to a computer - Change it to for
your computer.
Then we can plot just as above using the phylomatic() function
plot(tree, no.margin = TRUE)
Citing
To cite brranching in publications use:
Scott Chamberlain (2016). brranching: Fetch ‘Phylogenies’ from Many Sources. R package version 0.2.0. https://github.com/ropensci/brranching
License and bugs
- License: MIT
- Report bugs at our Github repo for rnoaa
