AntWeb tutorial
for v0.7.0
AntWeb is a repository that boasts a wealth of natural history data, digital images, and specimen records on ant species from a large community of museum curators.
Installation
install.packages("AntWeb")
or you can install the latest development version.
library("devtools")
install_github("ropensci/AntWeb")
Usage
Searching through the database
As with most of our packages, there are several ways to search through an API. In the case of AntWeb, you can search by a genus or full species name or by other taxonomic ranks like sub-phylum.
To obtain data on any taxonomic group, you can make a request using the aw_data() function. It’s possible to search easily by a taxonomic rank (e.g. a genus) or by passing a complete scientific name.
Searching by Genus
To get data on an ant genus found widely through Central and South America
library("AntWeb")
leaf_cutter_ants <- aw_data(genus = "acromyrmex")
leaf_cutter_ants$count
#> [1] 964
Searching by species
You can request data on any particular species
(acanthognathus_df <- aw_data(scientific_name = "acanthognathus brevicornis"))
#> [Total results on the server]: 4
#> [Args]:
#> genus = acanthognathus
#> species = brevicornis
#> [Dataset]: [4 x 16]
#> [Data preview] :
#> url
#> 1 http://antweb.org/api/v2/?occurrenceId=CAS:ANTWEB:casent0280684
#> 2 http://antweb.org/api/v2/?occurrenceId=CAS:ANTWEB:casent0637708
#> catalogNumber family subfamily genus specificEpithet
#> 1 casent0280684 formicidae myrmicinae Acanthognathus brevicornis
#> 2 casent0637708 formicidae myrmicinae Acanthognathus brevicornis
#> scientific_name typeStatus stateProvince country
#> 1 acanthognathus brevicornis Colombia
#> 2 acanthognathus brevicornis Madre de Dios Peru
#> dateIdentified habitat
#> 1
#> 2 2013-09-12 Bamboo forest ex sifted leaf litter
#> minimumElevationInMeters geojson.type decimal_latitude decimal_longitude
#> 1 NA <NA> <NA> <NA>
#> 2 252 point -13.14142 -69.623
You can also limit queries to observation records that have been geoferenced
(acanthognathus_df_geo <- aw_data(genus = "acanthognathus", species = "brevicornis", georeferenced = TRUE))
#> [Total results on the server]: 3
#> [Args]:
#> genus = acanthognathus
#> species = brevicornis
#> georeferenced = TRUE
#> [Dataset]: [3 x 16]
#> [Data preview] :
#> url
#> 1 http://antweb.org/api/v2/?occurrenceId=CAS:ANTWEB:casent0637708
#> 2 http://antweb.org/api/v2/?occurrenceId=CAS:ANTWEB:casent0914659
#> catalogNumber family subfamily genus specificEpithet
#> 1 casent0637708 formicidae myrmicinae Acanthognathus brevicornis
#> 2 casent0914659 formicidae myrmicinae Acanthognathus brevicornis
#> scientific_name typeStatus stateProvince country
#> 1 acanthognathus brevicornis Madre de Dios Peru
#> 2 acanthognathus brevicornis Darien Panama
#> dateIdentified habitat
#> 1 2013-09-12 Bamboo forest ex sifted leaf litter
#> 2
#> minimumElevationInMeters geojson.type decimal_latitude decimal_longitude
#> 1 252 point -13.14142 -69.623
#> 2 500 point 7.71667 -77.7
It’s also possible to search for records around any location by specifying a search radius.
This will search for data on a 2 km radius around that latitude/longitude
data_by_loc <- aw_coords(coord = "37.76,-122.45", r = 2)
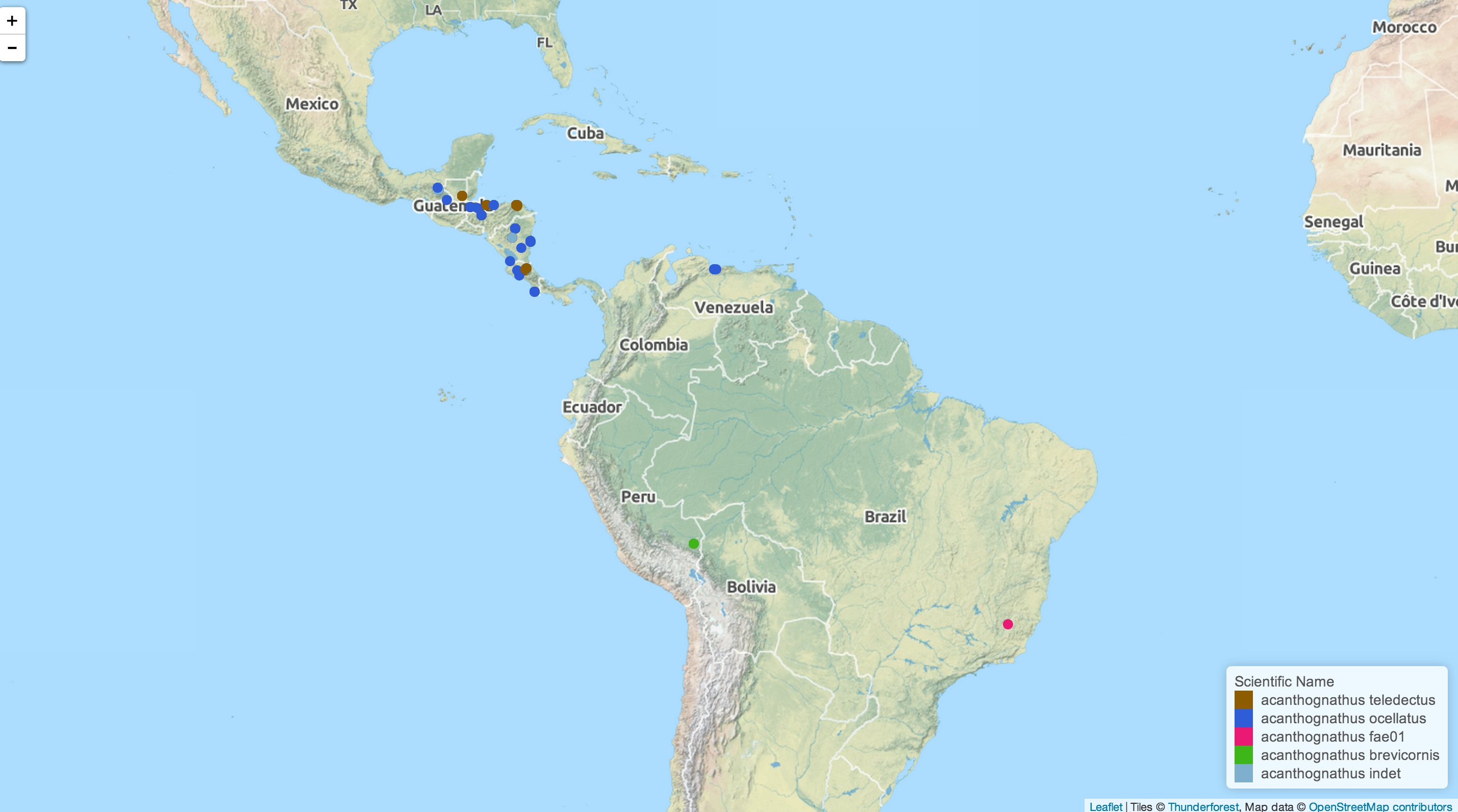
Mapping ant specimen data
As with the previous ecoengine package, you can also visualize location data for any set of species. Adding georeferenced = TRUE to a data retrieval call will filter out any data points without location information. Once retrieved the data are mapped with the open source Leaflet.js and pushed to your default browser. Maps and associated geoJSON files are also saved to a location specified (or defaults to your /tmp folder). This feature is only available on the development version on GitHub (0.5.2 or greater; see above on how to install) and will be available from CRAN in version 0.6
acd <- aw_data(genus = "acanthognathus")
aw_map(acd)
Citing
To cite AntWeb in publications use:
‘Karthik Ram’ (2014). AntWeb: programmatic interface to the AntWeb. R package version 0.6.1. http://CRAN.R-project.org/package=AntWeb
License and bugs
- License: CC0
- Report bugs at our Github repo for AntWeb
